Work with Multi-Band Rasters (Advance)
Last updated on 2024-03-12 | Edit this page
Estimated time: 60 minutes
WARNING
Warning in
download.file("https://www.naturalearthdata.com/http//www.naturalearthdata.com/download/110m/physical/ne_110m_graticules_all.zip",
: cannot open URL
'https://www.naturalearthdata.com/http//www.naturalearthdata.com/download/110m/physical/ne_110m_graticules_all.zip':
HTTP status was '500 Internal Server Error'ERROR
Error in download.file("https://www.naturalearthdata.com/http//www.naturalearthdata.com/download/110m/physical/ne_110m_graticules_all.zip", : cannot open URL 'https://www.naturalearthdata.com/http//www.naturalearthdata.com/download/110m/physical/ne_110m_graticules_all.zip'Overview
Questions
- How can I visualize individual and multiple bands in a raster object?
Objectives
- Identify a single vs. a multi-band raster file.
- Import multi-band rasters into R using the
terrapackage. - Plot multi-band color image rasters in R using the
ggplotpackage.
Things You’ll Need To Complete This Episode
See the lesson homepage for detailed information about the software, data, and other prerequisites you will need to work through the examples in this episode.
We introduced multi-band raster data in an earlier lesson. This episode explores how to import and plot a multi-band raster in R.
Getting Started with Multi-Band Data in R
In this episode, the multi-band data that we are working with is imagery collected using the NEON Airborne Observation Platform high resolution camera over the NEON Harvard Forest field site. Each RGB image is a 3-band raster. The same steps would apply to working with a multi-spectral image with 4 or more bands - like Landsat imagery.
By using the rast() function along with the
lyrs parameter, we can read specific raster bands (i.e. the
first one); omitting this parater would read instead all bands.
R
RGB_band1_HARV <-
rast("data/NEON-DS-Airborne-Remote-Sensing/HARV/RGB_Imagery/HARV_RGB_Ortho.tif",
lyrs = 1)
We need to convert this data to a data frame in order to plot it with
ggplot.
R
RGB_band1_HARV_df <- as.data.frame(RGB_band1_HARV, xy = TRUE)
R
ggplot() +
geom_raster(data = RGB_band1_HARV_df,
aes(x = x, y = y, alpha = HARV_RGB_Ortho_1)) +
coord_quickmap()
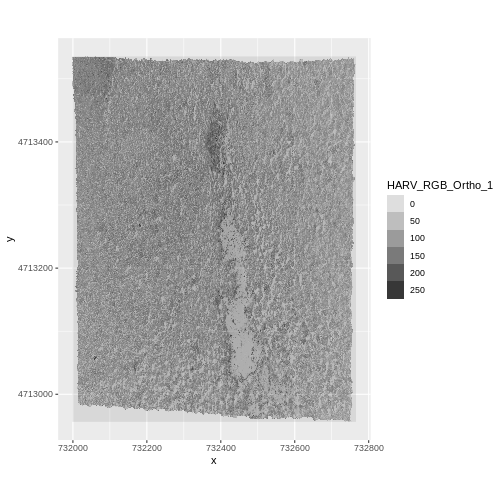
R
RGB_band1_HARV
OUTPUT
class : SpatRaster
dimensions : 2317, 3073, 1 (nrow, ncol, nlyr)
resolution : 0.25, 0.25 (x, y)
extent : 731998.5, 732766.8, 4712956, 4713536 (xmin, xmax, ymin, ymax)
coord. ref. : WGS 84 / UTM zone 18N (EPSG:32618)
source : HARV_RGB_Ortho.tif
name : HARV_RGB_Ortho_1
min value : 0
max value : 255 Notice that when we look at the attributes of this band, we see:
dimensions : 2317, 3073, 1 (nrow, ncol, nlyr)
This is R telling us that we read only one its bands.
Image Raster Data Values
As we saw in the previous exercise, this raster contains values between 0 and 255. These values represent degrees of brightness associated with the image band. In the case of a RGB image (red, green and blue), band 1 is the red band. When we plot the red band, larger numbers (towards 255) represent pixels with more red in them (a strong red reflection). Smaller numbers (towards 0) represent pixels with less red in them (less red was reflected). To plot an RGB image, we mix red + green + blue values into one single color to create a full color image - similar to the color image a digital camera creates.
Import A Specific Band
We can use the rast() function to import specific bands
in our raster object by specifying which band we want with
lyrs = N (N represents the band number we want to work
with). To import the green band, we would use lyrs = 2.
R
RGB_band2_HARV <-
rast("data/NEON-DS-Airborne-Remote-Sensing/HARV/RGB_Imagery/HARV_RGB_Ortho.tif",
lyrs = 2)
We can convert this data to a data frame and plot the same way we plotted the red band:
R
RGB_band2_HARV_df <- as.data.frame(RGB_band2_HARV, xy = TRUE)
R
ggplot() +
geom_raster(data = RGB_band2_HARV_df,
aes(x = x, y = y, alpha = HARV_RGB_Ortho_2)) +
coord_equal()

We’d expect a brighter value for the forest in band 2 (green) than in band 1 (red) because the leaves on trees of most often appear “green” - healthy leaves reflect MORE green light than red light.
Raster Stacks in R
Next, we will work with all three image bands (red, green and blue) as an R raster object. We will then plot a 3-band composite, or full color, image.
To bring in all bands of a multi-band raster, we use
therast() function.
R
RGB_stack_HARV <-
rast("data/NEON-DS-Airborne-Remote-Sensing/HARV/RGB_Imagery/HARV_RGB_Ortho.tif")
Let’s preview the attributes of our stack object:
R
RGB_stack_HARV
OUTPUT
class : SpatRaster
dimensions : 2317, 3073, 3 (nrow, ncol, nlyr)
resolution : 0.25, 0.25 (x, y)
extent : 731998.5, 732766.8, 4712956, 4713536 (xmin, xmax, ymin, ymax)
coord. ref. : WGS 84 / UTM zone 18N (EPSG:32618)
source : HARV_RGB_Ortho.tif
names : HARV_RGB_Ortho_1, HARV_RGB_Ortho_2, HARV_RGB_Ortho_3
min values : 0, 0, 0
max values : 255, 255, 255 We can view the attributes of each band in the stack in a single output. For example, if we had hundreds of bands, we could specify which band we’d like to view attributes for using an index value:
R
RGB_stack_HARV[[2]]
OUTPUT
class : SpatRaster
dimensions : 2317, 3073, 1 (nrow, ncol, nlyr)
resolution : 0.25, 0.25 (x, y)
extent : 731998.5, 732766.8, 4712956, 4713536 (xmin, xmax, ymin, ymax)
coord. ref. : WGS 84 / UTM zone 18N (EPSG:32618)
source : HARV_RGB_Ortho.tif
name : HARV_RGB_Ortho_2
min value : 0
max value : 255 We can also use the ggplot functions to plot the data in
any layer of our raster object. Remember, we need to convert to a data
frame first.
R
RGB_stack_HARV_df <- as.data.frame(RGB_stack_HARV, xy = TRUE)
Each band in our RasterStack gets its own column in the data frame. Thus we have:
R
str(RGB_stack_HARV_df)
OUTPUT
'data.frame': 7120141 obs. of 5 variables:
$ x : num 731999 731999 731999 731999 732000 ...
$ y : num 4713535 4713535 4713535 4713535 4713535 ...
$ HARV_RGB_Ortho_1: num 0 2 6 0 16 0 0 6 1 5 ...
$ HARV_RGB_Ortho_2: num 1 0 9 0 5 0 4 2 1 0 ...
$ HARV_RGB_Ortho_3: num 0 10 1 0 17 0 4 0 0 7 ...Let’s create a histogram of the first band:
R
ggplot() +
geom_histogram(data = RGB_stack_HARV_df, aes(HARV_RGB_Ortho_1))
OUTPUT
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.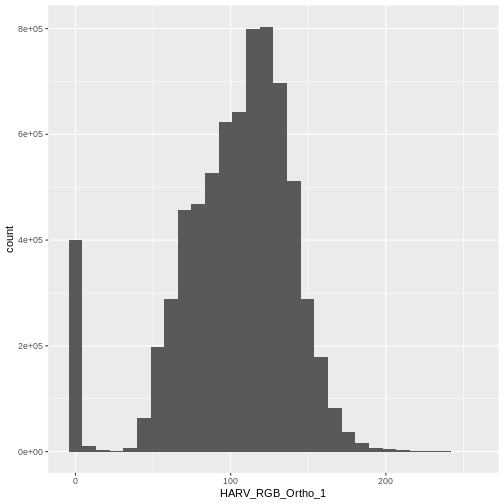
And a raster plot of the second band:
R
ggplot() +
geom_raster(data = RGB_stack_HARV_df,
aes(x = x, y = y, alpha = HARV_RGB_Ortho_2)) +
coord_quickmap()

We can access any individual band in the same way.
Create A Three Band Image
To render a final three band, colored image in R, we use the
plotRGB() function.
This function allows us to:
- Identify what bands we want to render in the red, green and blue
regions. The
plotRGB()function defaults to a 1=red, 2=green, and 3=blue band order. However, you can define what bands you’d like to plot manually. Manual definition of bands is useful if you have, for example a near-infrared band and want to create a color infrared image. - Adjust the
stretchof the image to increase or decrease contrast.
Let’s plot our 3-band image. Note that we can use the
plotRGB() function directly with our RasterStack object (we
don’t need a dataframe as this function isn’t part of the
ggplot2 package).
R
plotRGB(RGB_stack_HARV,
r = 1, g = 2, b = 3)

The image above looks pretty good. We can explore whether applying a
stretch to the image might improve clarity and contrast using
stretch="lin" or stretch="hist".
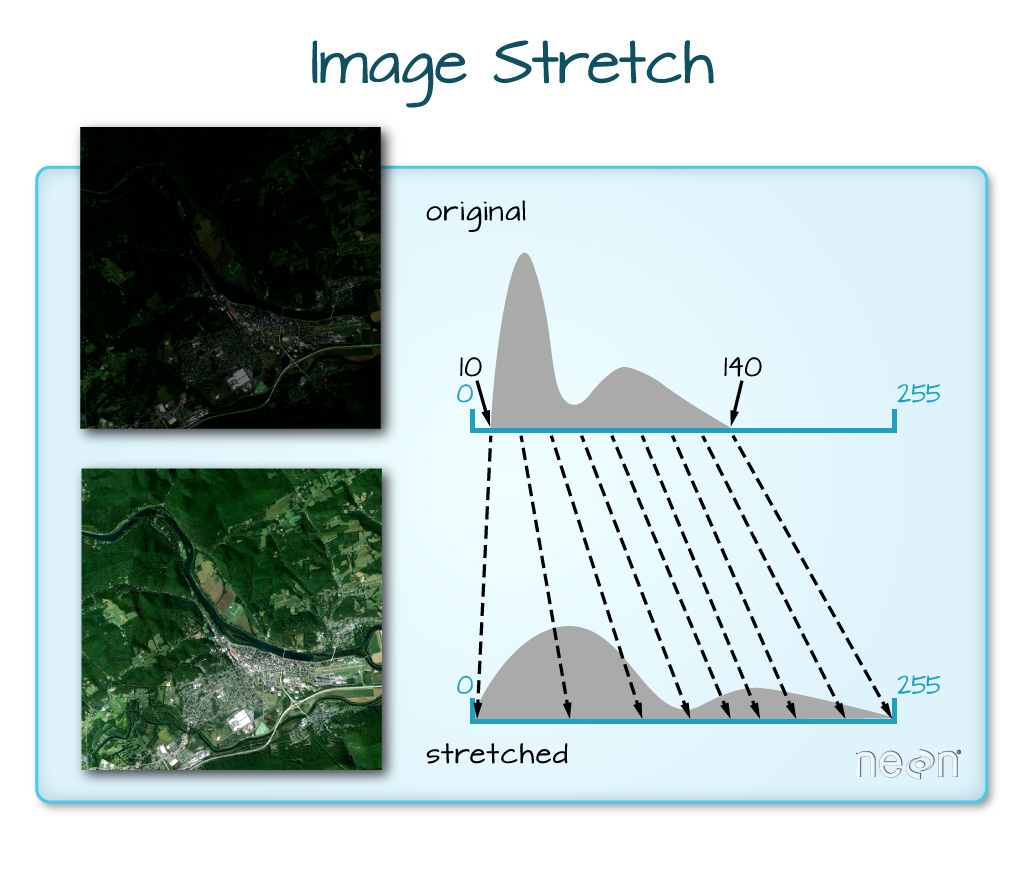
When the range of pixel brightness values is closer to 0, a darker image is rendered by default. We can stretch the values to extend to the full 0-255 range of potential values to increase the visual contrast of the image.
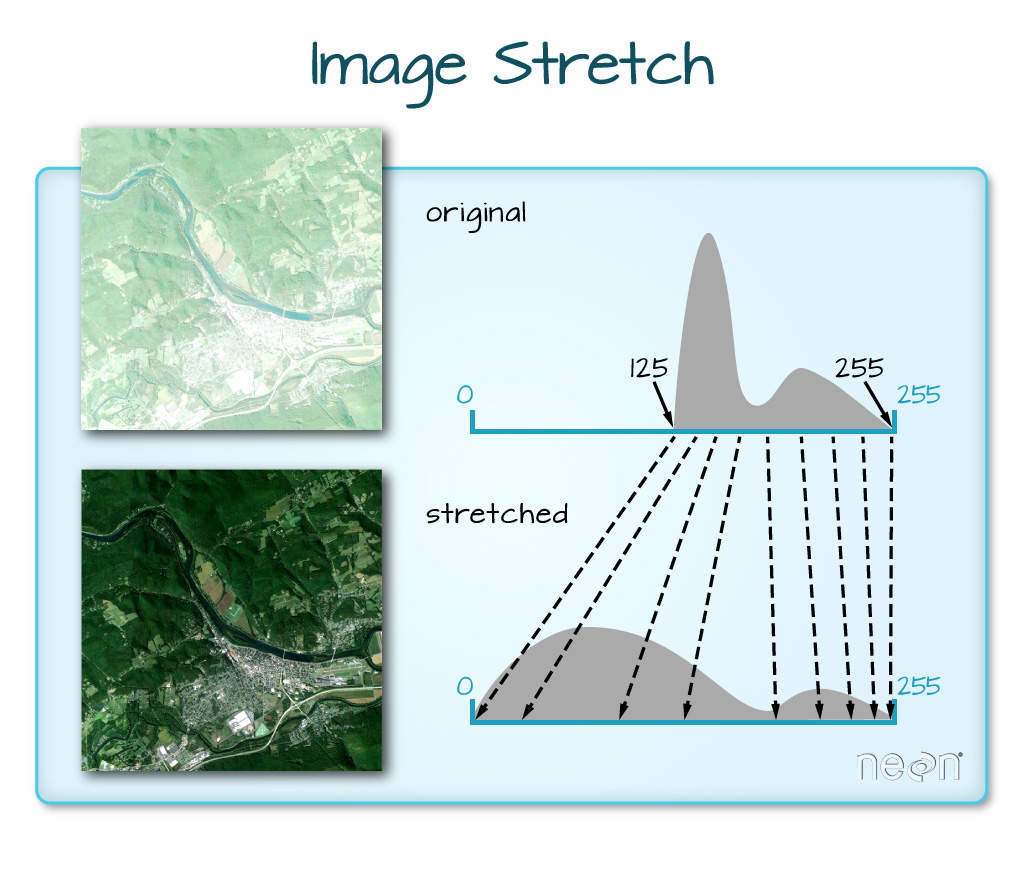
When the range of pixel brightness values is closer to 255, a lighter image is rendered by default. We can stretch the values to extend to the full 0-255 range of potential values to increase the visual contrast of the image.
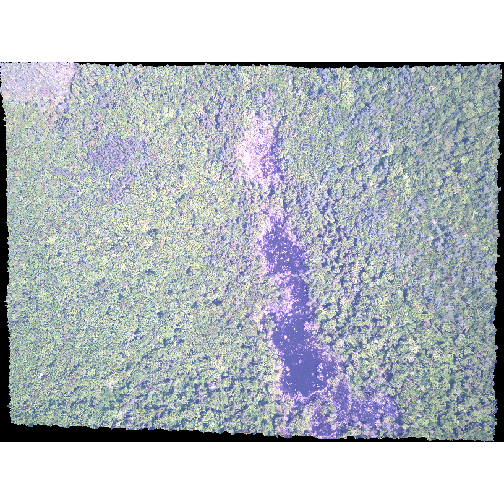
R
plotRGB(RGB_stack_HARV,
r = 1, g = 2, b = 3,
scale = 800,
stretch = "lin")
R
plotRGB(RGB_stack_HARV,
r = 1, g = 2, b = 3,
scale = 800,
stretch = "hist")

In this case, the stretch doesn’t enhance the contrast our image significantly given the distribution of reflectance (or brightness) values is distributed well between 0 and 255.
Challenge - NoData Values
Let’s explore what happens with NoData values when working with
RasterStack objects and using the plotRGB() function. We
will use the HARV_Ortho_wNA.tif GeoTIFF file in the
NEON-DS-Airborne-Remote-Sensing/HARVRGB_Imagery/
directory.
- View the files attributes. Are there
NoDatavalues assigned for this file? - If so, what is the
NoDataValue? - How many bands does it have?
- Load the multi-band raster file into R.
- Plot the object as a true color image.
- What happened to the black edges in the data?
- What does this tell us about the difference in the data structure
between
HARV_Ortho_wNA.tifandHARV_RGB_Ortho.tif(R objectRGB_stack). How can you check?
- First we use the
describe()function to view the data attributes.
R
describe("data/NEON-DS-Airborne-Remote-Sensing/HARV/RGB_Imagery/HARV_Ortho_wNA.tif")
OUTPUT
[1] "Driver: GTiff/GeoTIFF"
[2] "Files: data/NEON-DS-Airborne-Remote-Sensing/HARV/RGB_Imagery/HARV_Ortho_wNA.tif"
[3] "Size is 3073, 2317"
[4] "Coordinate System is:"
[5] "PROJCRS[\"WGS 84 / UTM zone 18N\","
[6] " BASEGEOGCRS[\"WGS 84\","
[7] " DATUM[\"World Geodetic System 1984\","
[8] " ELLIPSOID[\"WGS 84\",6378137,298.257223563,"
[9] " LENGTHUNIT[\"metre\",1]]],"
[10] " PRIMEM[\"Greenwich\",0,"
[11] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
[12] " ID[\"EPSG\",4326]],"
[13] " CONVERSION[\"UTM zone 18N\","
[14] " METHOD[\"Transverse Mercator\","
[15] " ID[\"EPSG\",9807]],"
[16] " PARAMETER[\"Latitude of natural origin\",0,"
[17] " ANGLEUNIT[\"degree\",0.0174532925199433],"
[18] " ID[\"EPSG\",8801]],"
[19] " PARAMETER[\"Longitude of natural origin\",-75,"
[20] " ANGLEUNIT[\"degree\",0.0174532925199433],"
[21] " ID[\"EPSG\",8802]],"
[22] " PARAMETER[\"Scale factor at natural origin\",0.9996,"
[23] " SCALEUNIT[\"unity\",1],"
[24] " ID[\"EPSG\",8805]],"
[25] " PARAMETER[\"False easting\",500000,"
[26] " LENGTHUNIT[\"metre\",1],"
[27] " ID[\"EPSG\",8806]],"
[28] " PARAMETER[\"False northing\",0,"
[29] " LENGTHUNIT[\"metre\",1],"
[30] " ID[\"EPSG\",8807]]],"
[31] " CS[Cartesian,2],"
[32] " AXIS[\"(E)\",east,"
[33] " ORDER[1],"
[34] " LENGTHUNIT[\"metre\",1]],"
[35] " AXIS[\"(N)\",north,"
[36] " ORDER[2],"
[37] " LENGTHUNIT[\"metre\",1]],"
[38] " USAGE["
[39] " SCOPE[\"Engineering survey, topographic mapping.\"],"
[40] " AREA[\"Between 78°W and 72°W, northern hemisphere between equator and 84°N, onshore and offshore. Bahamas. Canada - Nunavut; Ontario; Quebec. Colombia. Cuba. Ecuador. Greenland. Haiti. Jamica. Panama. Turks and Caicos Islands. United States (USA). Venezuela.\"],"
[41] " BBOX[0,-78,84,-72]],"
[42] " ID[\"EPSG\",32618]]"
[43] "Data axis to CRS axis mapping: 1,2"
[44] "Origin = (731998.500000000000000,4713535.500000000000000)"
[45] "Pixel Size = (0.250000000000000,-0.250000000000000)"
[46] "Metadata:"
[47] " AREA_OR_POINT=Area"
[48] "Image Structure Metadata:"
[49] " COMPRESSION=LZW"
[50] " INTERLEAVE=PIXEL"
[51] "Corner Coordinates:"
[52] "Upper Left ( 731998.500, 4713535.500) ( 72d10'29.27\"W, 42d32'21.80\"N)"
[53] "Lower Left ( 731998.500, 4712956.250) ( 72d10'30.11\"W, 42d32' 3.04\"N)"
[54] "Upper Right ( 732766.750, 4713535.500) ( 72d 9'55.63\"W, 42d32'20.97\"N)"
[55] "Lower Right ( 732766.750, 4712956.250) ( 72d 9'56.48\"W, 42d32' 2.21\"N)"
[56] "Center ( 732382.625, 4713245.875) ( 72d10'12.87\"W, 42d32'12.00\"N)"
[57] "Band 1 Block=3073x1 Type=Float64, ColorInterp=Gray"
[58] " Min=0.000 Max=255.000 "
[59] " Minimum=0.000, Maximum=255.000, Mean=107.837, StdDev=30.019"
[60] " NoData Value=-9999"
[61] " Metadata:"
[62] " STATISTICS_MAXIMUM=255"
[63] " STATISTICS_MEAN=107.83651227531"
[64] " STATISTICS_MINIMUM=0"
[65] " STATISTICS_STDDEV=30.019177549096"
[66] "Band 2 Block=3073x1 Type=Float64, ColorInterp=Undefined"
[67] " Min=0.000 Max=255.000 "
[68] " Minimum=0.000, Maximum=255.000, Mean=130.096, StdDev=32.002"
[69] " NoData Value=-9999"
[70] " Metadata:"
[71] " STATISTICS_MAXIMUM=255"
[72] " STATISTICS_MEAN=130.09595363812"
[73] " STATISTICS_MINIMUM=0"
[74] " STATISTICS_STDDEV=32.001675868273"
[75] "Band 3 Block=3073x1 Type=Float64, ColorInterp=Undefined"
[76] " Min=0.000 Max=255.000 "
[77] " Minimum=0.000, Maximum=255.000, Mean=95.760, StdDev=16.577"
[78] " NoData Value=-9999"
[79] " Metadata:"
[80] " STATISTICS_MAXIMUM=255"
[81] " STATISTICS_MEAN=95.759787935476"
[82] " STATISTICS_MINIMUM=0"
[83] " STATISTICS_STDDEV=16.577042076977" From the output above, we see that there are
NoDatavalues and they are assigned the value of -9999.The data has three bands.
To read in the file, we will use the
rast()function:
R
HARV_NA <-
rast("data/NEON-DS-Airborne-Remote-Sensing/HARV/RGB_Imagery/HARV_Ortho_wNA.tif")
- We can plot the data with the
plotRGB()function:
R
plotRGB(HARV_NA,
r = 1, g = 2, b = 3)

The black edges are not plotted.
Both data sets have
NoDatavalues, however, in the RGB_stack the NoData value is not defined in the tiff tags, thus R renders them as black as the reflectance values are 0. The black edges in the other file are defined as -9999 and R renders them as NA.
R
describe("data/NEON-DS-Airborne-Remote-Sensing/HARV/RGB_Imagery/HARV_RGB_Ortho.tif")
OUTPUT
[1] "Driver: GTiff/GeoTIFF"
[2] "Files: data/NEON-DS-Airborne-Remote-Sensing/HARV/RGB_Imagery/HARV_RGB_Ortho.tif"
[3] "Size is 3073, 2317"
[4] "Coordinate System is:"
[5] "PROJCRS[\"WGS 84 / UTM zone 18N\","
[6] " BASEGEOGCRS[\"WGS 84\","
[7] " DATUM[\"World Geodetic System 1984\","
[8] " ELLIPSOID[\"WGS 84\",6378137,298.257223563,"
[9] " LENGTHUNIT[\"metre\",1]]],"
[10] " PRIMEM[\"Greenwich\",0,"
[11] " ANGLEUNIT[\"degree\",0.0174532925199433]],"
[12] " ID[\"EPSG\",4326]],"
[13] " CONVERSION[\"UTM zone 18N\","
[14] " METHOD[\"Transverse Mercator\","
[15] " ID[\"EPSG\",9807]],"
[16] " PARAMETER[\"Latitude of natural origin\",0,"
[17] " ANGLEUNIT[\"degree\",0.0174532925199433],"
[18] " ID[\"EPSG\",8801]],"
[19] " PARAMETER[\"Longitude of natural origin\",-75,"
[20] " ANGLEUNIT[\"degree\",0.0174532925199433],"
[21] " ID[\"EPSG\",8802]],"
[22] " PARAMETER[\"Scale factor at natural origin\",0.9996,"
[23] " SCALEUNIT[\"unity\",1],"
[24] " ID[\"EPSG\",8805]],"
[25] " PARAMETER[\"False easting\",500000,"
[26] " LENGTHUNIT[\"metre\",1],"
[27] " ID[\"EPSG\",8806]],"
[28] " PARAMETER[\"False northing\",0,"
[29] " LENGTHUNIT[\"metre\",1],"
[30] " ID[\"EPSG\",8807]]],"
[31] " CS[Cartesian,2],"
[32] " AXIS[\"(E)\",east,"
[33] " ORDER[1],"
[34] " LENGTHUNIT[\"metre\",1]],"
[35] " AXIS[\"(N)\",north,"
[36] " ORDER[2],"
[37] " LENGTHUNIT[\"metre\",1]],"
[38] " USAGE["
[39] " SCOPE[\"Engineering survey, topographic mapping.\"],"
[40] " AREA[\"Between 78°W and 72°W, northern hemisphere between equator and 84°N, onshore and offshore. Bahamas. Canada - Nunavut; Ontario; Quebec. Colombia. Cuba. Ecuador. Greenland. Haiti. Jamica. Panama. Turks and Caicos Islands. United States (USA). Venezuela.\"],"
[41] " BBOX[0,-78,84,-72]],"
[42] " ID[\"EPSG\",32618]]"
[43] "Data axis to CRS axis mapping: 1,2"
[44] "Origin = (731998.500000000000000,4713535.500000000000000)"
[45] "Pixel Size = (0.250000000000000,-0.250000000000000)"
[46] "Metadata:"
[47] " AREA_OR_POINT=Area"
[48] "Image Structure Metadata:"
[49] " COMPRESSION=LZW"
[50] " INTERLEAVE=PIXEL"
[51] "Corner Coordinates:"
[52] "Upper Left ( 731998.500, 4713535.500) ( 72d10'29.27\"W, 42d32'21.80\"N)"
[53] "Lower Left ( 731998.500, 4712956.250) ( 72d10'30.11\"W, 42d32' 3.04\"N)"
[54] "Upper Right ( 732766.750, 4713535.500) ( 72d 9'55.63\"W, 42d32'20.97\"N)"
[55] "Lower Right ( 732766.750, 4712956.250) ( 72d 9'56.48\"W, 42d32' 2.21\"N)"
[56] "Center ( 732382.625, 4713245.875) ( 72d10'12.87\"W, 42d32'12.00\"N)"
[57] "Band 1 Block=3073x1 Type=Float64, ColorInterp=Gray"
[58] " Min=0.000 Max=255.000 "
[59] " Minimum=0.000, Maximum=255.000, Mean=nan, StdDev=nan"
[60] " NoData Value=-1.69999999999999994e+308"
[61] " Metadata:"
[62] " STATISTICS_MAXIMUM=255"
[63] " STATISTICS_MEAN=nan"
[64] " STATISTICS_MINIMUM=0"
[65] " STATISTICS_STDDEV=nan"
[66] "Band 2 Block=3073x1 Type=Float64, ColorInterp=Undefined"
[67] " Min=0.000 Max=255.000 "
[68] " Minimum=0.000, Maximum=255.000, Mean=nan, StdDev=nan"
[69] " NoData Value=-1.69999999999999994e+308"
[70] " Metadata:"
[71] " STATISTICS_MAXIMUM=255"
[72] " STATISTICS_MEAN=nan"
[73] " STATISTICS_MINIMUM=0"
[74] " STATISTICS_STDDEV=nan"
[75] "Band 3 Block=3073x1 Type=Float64, ColorInterp=Undefined"
[76] " Min=0.000 Max=255.000 "
[77] " Minimum=0.000, Maximum=255.000, Mean=nan, StdDev=nan"
[78] " NoData Value=-1.69999999999999994e+308"
[79] " Metadata:"
[80] " STATISTICS_MAXIMUM=255"
[81] " STATISTICS_MEAN=nan"
[82] " STATISTICS_MINIMUM=0"
[83] " STATISTICS_STDDEV=nan" Data Tip
We can create a raster object from several, individual single-band GeoTIFFs too. We will do this in a later episode, Raster Time Series Data in R.
SpatRaster in R
The R SpatRaster object type can handle rasters with multiple bands. The SpatRaster only holds parameters that describe the properties of raster data that is located somewhere on our computer.
A SpatRasterDataset object can hold references to sub-datasets, that is, SpatRaster objects. In most cases, we can work with a SpatRaster in the same way we might work with a SpatRasterDataset.
We can build a SpatRasterDataset using a SpatRaster or a list of SpatRaster:
R
RGB_sds_HARV <- sds(RGB_stack_HARV)
RGB_sds_HARV <- sds(list(RGB_stack_HARV, RGB_stack_HARV))
We can retrieve the SpatRaster objects from a SpatRasterDataset using subsetting:
R
RGB_sds_HARV[[1]]
OUTPUT
class : SpatRaster
dimensions : 2317, 3073, 3 (nrow, ncol, nlyr)
resolution : 0.25, 0.25 (x, y)
extent : 731998.5, 732766.8, 4712956, 4713536 (xmin, xmax, ymin, ymax)
coord. ref. : WGS 84 / UTM zone 18N (EPSG:32618)
source : HARV_RGB_Ortho.tif
names : HARV_RGB_Ortho_1, HARV_RGB_Ortho_2, HARV_RGB_Ortho_3
min values : 0, 0, 0
max values : 255, 255, 255 R
RGB_sds_HARV[[2]]
OUTPUT
class : SpatRaster
dimensions : 2317, 3073, 3 (nrow, ncol, nlyr)
resolution : 0.25, 0.25 (x, y)
extent : 731998.5, 732766.8, 4712956, 4713536 (xmin, xmax, ymin, ymax)
coord. ref. : WGS 84 / UTM zone 18N (EPSG:32618)
source : HARV_RGB_Ortho.tif
names : HARV_RGB_Ortho_1, HARV_RGB_Ortho_2, HARV_RGB_Ortho_3
min values : 0, 0, 0
max values : 255, 255, 255 Challenge: What Functions Can Be Used on an R Object of a particular class?
We can view various functions (or methods) available to use on an R
object with methods(class=class(objectNameHere)). Use this
to figure out:
- What methods can be used on the
RGB_stack_HARVobject? - What methods can be used on a single band within
RGB_stack_HARV? - Why do you think there isn’t a difference?
- We can see a list of all of the methods available for our RasterStack object:
R
methods(class=class(RGB_stack_HARV))
OUTPUT
[1] ! [ [[
[4] [[<- [<- %in%
[7] $ $<- activeCat
[10] activeCat<- add<- addCats
[13] adjacent aggregate align
[16] all.equal allNA animate
[19] anyNA app approximate
[22] area Arith as.array
[25] as.bool as.character as.contour
[28] as.data.frame as.factor as.int
[31] as.integer as.lines as.list
[34] as.logical as.matrix as.numeric
[37] as.points as.polygons as.raster
[40] atan_2 atan2 autocor
[43] barplot blocks boundaries
[46] boxplot buffer c
[49] catalyze categories cats
[52] cellFromRowCol cellFromRowColCombine cellFromXY
[55] cells cellSize clamp_ts
[58] clamp classify click
[61] coerce colFromCell colFromX
[64] colorize coltab coltab<-
[67] Compare compare compareGeom
[70] concats contour costDist
[73] countNA cover crds
[76] crop crosstab crs
[79] crs<- datatype deepcopy
[82] density depth depth<-
[85] diff dim dim<-
[88] direction disagg distance
[91] droplevels expanse ext
[94] ext<- extend extract
[97] fillTime flip focal
[100] focal3D focalCor focalCpp
[103] focalPairs focalReg focalValues
[106] freq global gridDist
[109] gridDistance has.colors has.RGB
[112] hasMinMax hasValues head
[115] hist ifel image
[118] init inMemory inset
[121] interpIDW interpNear interpolate
[124] intersect is.bool is.factor
[127] is.finite is.infinite is.int
[130] is.lonlat is.na is.nan
[133] is.related isFALSE isTRUE
[136] lapp layerCor levels
[139] levels<- linearUnits lines
[142] log Logic logic
[145] longnames longnames<- makeTiles
[148] mask match math
[151] Math Math2 mean
[154] median merge meta
[157] minmax modal mosaic
[160] NAflag NAflag<- names
[163] names<- ncell ncol
[166] ncol<- nlyr nlyr<-
[169] noNA not.na nrow
[172] nrow<- nsrc origin
[175] origin<- pairs panel
[178] patches persp plet
[181] plot plotRGB points
[184] polys predict project
[187] quantile rangeFill rapp
[190] rast rasterize rasterizeGeom
[193] rasterizeWin rcl readStart
[196] readStop readValues rectify
[199] regress relate rep
[202] res res<- resample
[205] rescale rev RGB
[208] RGB<- roll rotate
[211] rowColCombine rowColFromCell rowFromCell
[214] rowFromY sapp saveRDS
[217] scale scoff scoff<-
[220] sds segregate sel
[223] selectHighest selectRange serialize
[226] set.cats set.crs set.ext
[229] set.names set.RGB set.values
[232] setMinMax setValues shift
[235] show sieve size
[238] sort sources spatSample
[241] split sprc stdev
[244] str stretch subset
[247] subst summary Summary
[250] t tail tapp
[253] terrain text tighten
[256] time time<- timeInfo
[259] trans trim unique
[262] units units<- update
[265] values values<- varnames
[268] varnames<- viewshed weighted.mean
[271] where.max where.min which.lyr
[274] which.max which.min window
[277] window<- wrap wrapCache
[280] writeCDF writeRaster writeStart
[283] writeStop writeValues xFromCell
[286] xFromCol xmax xmax<-
[289] xmin xmin<- xres
[292] xyFromCell yFromCell yFromRow
[295] ymax ymax<- ymin
[298] ymin<- yres zonal
[301] zoom
see '?methods' for accessing help and source code- And compare that with the methods available for a single band:
R
methods(class=class(RGB_stack_HARV[[1]]))
OUTPUT
[1] ! [ [[
[4] [[<- [<- %in%
[7] $ $<- activeCat
[10] activeCat<- add<- addCats
[13] adjacent aggregate align
[16] all.equal allNA animate
[19] anyNA app approximate
[22] area Arith as.array
[25] as.bool as.character as.contour
[28] as.data.frame as.factor as.int
[31] as.integer as.lines as.list
[34] as.logical as.matrix as.numeric
[37] as.points as.polygons as.raster
[40] atan_2 atan2 autocor
[43] barplot blocks boundaries
[46] boxplot buffer c
[49] catalyze categories cats
[52] cellFromRowCol cellFromRowColCombine cellFromXY
[55] cells cellSize clamp_ts
[58] clamp classify click
[61] coerce colFromCell colFromX
[64] colorize coltab coltab<-
[67] Compare compare compareGeom
[70] concats contour costDist
[73] countNA cover crds
[76] crop crosstab crs
[79] crs<- datatype deepcopy
[82] density depth depth<-
[85] diff dim dim<-
[88] direction disagg distance
[91] droplevels expanse ext
[94] ext<- extend extract
[97] fillTime flip focal
[100] focal3D focalCor focalCpp
[103] focalPairs focalReg focalValues
[106] freq global gridDist
[109] gridDistance has.colors has.RGB
[112] hasMinMax hasValues head
[115] hist ifel image
[118] init inMemory inset
[121] interpIDW interpNear interpolate
[124] intersect is.bool is.factor
[127] is.finite is.infinite is.int
[130] is.lonlat is.na is.nan
[133] is.related isFALSE isTRUE
[136] lapp layerCor levels
[139] levels<- linearUnits lines
[142] log Logic logic
[145] longnames longnames<- makeTiles
[148] mask match math
[151] Math Math2 mean
[154] median merge meta
[157] minmax modal mosaic
[160] NAflag NAflag<- names
[163] names<- ncell ncol
[166] ncol<- nlyr nlyr<-
[169] noNA not.na nrow
[172] nrow<- nsrc origin
[175] origin<- pairs panel
[178] patches persp plet
[181] plot plotRGB points
[184] polys predict project
[187] quantile rangeFill rapp
[190] rast rasterize rasterizeGeom
[193] rasterizeWin rcl readStart
[196] readStop readValues rectify
[199] regress relate rep
[202] res res<- resample
[205] rescale rev RGB
[208] RGB<- roll rotate
[211] rowColCombine rowColFromCell rowFromCell
[214] rowFromY sapp saveRDS
[217] scale scoff scoff<-
[220] sds segregate sel
[223] selectHighest selectRange serialize
[226] set.cats set.crs set.ext
[229] set.names set.RGB set.values
[232] setMinMax setValues shift
[235] show sieve size
[238] sort sources spatSample
[241] split sprc stdev
[244] str stretch subset
[247] subst summary Summary
[250] t tail tapp
[253] terrain text tighten
[256] time time<- timeInfo
[259] trans trim unique
[262] units units<- update
[265] values values<- varnames
[268] varnames<- viewshed weighted.mean
[271] where.max where.min which.lyr
[274] which.max which.min window
[277] window<- wrap wrapCache
[280] writeCDF writeRaster writeStart
[283] writeStop writeValues xFromCell
[286] xFromCol xmax xmax<-
[289] xmin xmin<- xres
[292] xyFromCell yFromCell yFromRow
[295] ymax ymax<- ymin
[298] ymin<- yres zonal
[301] zoom
see '?methods' for accessing help and source code- A SpatRaster is the same no matter its number of bands.
